
http://www.theguardian.com/environment/2016/mar/10/could-a-new-plastic-eating-bacteria-help-combat-this-pollution-scourge
Nature has begun to fight back against the vast piles of filth dumped into its soils, rivers and oceans by evolving a plastic-eating bacteria – the first known to science.
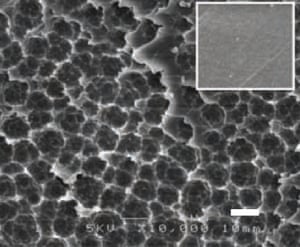
In a report published in the journal Science, a team of Japanese researchers described a species of bacteria that can break the molecular bonds of one of the world’s most-used plastics – polyethylene terephthalate, also known as PET or polyester.
The Japanese research team sifted through hundreds of samples of PET pollution before finding a colony of organisms using the plastic as a food source.
Further tests found the bacteria almost completely degraded low-quality plastic within six weeks. This was voracious when compared to other biological agents; including a related bacteria, leaf compost and a fungus enzyme recently found to have an appetite for PET.
“This is the first rigorous study – it appears to be very carefully done – that I have seen that shows plastic being hydrolyzed [broken down] by bacteria,” said Dr Tracy Mincer, a researcher at Woods Hole Oceanographic Institution.
The molecules that form PET are bonded very strongly, said Prof Uwe Bornscheuer in an accompanying comment piece in Science. “Until recently, no organisms were known to be able to decompose it.”
In a Gaian twist, initial genetic examination revealed the bacteria, namedIdeonella sakaiensis 201-F6, may have evolved enzymes specifically capable of breaking down PET in response to the accumulation of the plastic in the environment in the past 70 years.
Such rapid evolution was possible, said Enzo Palombo, a professor of microbiology at Swinburne University, given that microbes have an extraordinary ability to adapt to their surroundings. “If you put a bacteria in a situation where they’ve only got one food source to consume, over time they will adapt to do that,” he said.
“I think we are seeing how nature can surprise us and in the end the resiliency of nature itself,” added Mincer.
The bacteria took longer to eat away highly crystallised PET, which is used in plastic bottles. That means the enzymes and processes would need refinement before they could be useful for industrial recycling or pollution clean-up.
“It’s difficult to break down highly crystallised PET,” said Prof Kenji Miyamoto from Keio University, one of the authors of the study. “Our research results are just the initiation for the application. We have to work on so many issues needed for various applications. It takes a long time,” he said.
A third of all plastics end up in the environment and 8m tonnes end up in the ocean every year, creating vast accumulations of life-choking rubbish.
PET makes up almost one-sixth of the world’s annual plastic production of 311m tons. Despite PET being one of the more commonly recycled plastics, the World Economic Forum (WEF) reports that only just over half is ever collected for recycling and far less actually ends up being reused.
Advances in biodegradable plastics and recycling offer hope for the future, said Bornscheuer, “but [this] does not help to get rid of the plastics already in the environment”.
However the potential applications of the discovery remain unclear. The most obvious use would be as a biological agent in nature, said Palombo. Bacteria could be sprayed on the huge floating trash heaps building up in the oceans. This method is most notably employed to combat oil spills.
This particular bacteria would not be useful for this process as it only consumes PET, which is too dense to float on water. But Bornscheuer said the discovery could open the door to the discovery or manufacture of biological agents able to break down other plastics.
Palombo said the discovery suggested that other bacteria may have already evolved to do this job and simply needed to be found.
“I would not be surprised if samples of ocean plastics contained microbes that are happily growing on this material and could be isolated in the same manner,” he said.
But Mincer said breaking down ocean rubbish came with dangers of its own.Plastics often contain additives that can be toxic when released. WEF estimates that the 150m tonnes of plastic currently in the ocean contain roughly 23m tonnes of additives.
“Plastic debris may have been less toxic in the whole unhydrolyzed form where it would ultimately have been buried in the sediments on a geological timescale,” said Mincer.
Beyond dealing with the plastic already fouling up the environment, the bacteria could potentially be used in industrial recycling processes.
“Certainly, the use of these microbes or enzymes could play a role in remediation of plastic in a controlled reactor,” said Mincer.
Miyamoto’s team suggested that the environmentally-benign constituents left behind by the bacteria could be the same ones from which the plastic is formed. If this were true and a process could be developed to isolate them, Bornscheuer said: “This could provide huge savings in the production of new polymer without the need for petrol-based starting materials.” According to the WEF, 6% of global oil production is devoted to the production of plastics.
But the plastics industry said the potential for a new biological process to replace or augment the current mechanical recycling process was very small.
“PET is 100% recyclable,” said Mike Neal, the chairman of the Committee of PET Manufacturers in Europe. “I expect that a biodegradation system would require a similar engineering process to chemical depolymerisation and as such is unlikely to be economically viable,” he said.

